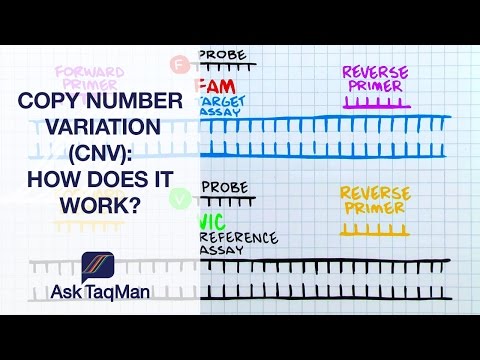
Q. How is gene copy number determined?
To measure DNA copy number, the amplicon should be located either within an exon or intron with sequences unique to that gene. A control gene with two copies should also be included.
Q. Can PCR detect CNV?
The advent of digital PCR (dPCR) now permits very high-resolution determination of CNV, often using smaller sample and reagent volumes. Droplet Digital™ PCR technology overcomes a number of inherent limitations of qPCR and microarray techniques for CNV analysis.
Table of Contents
- Q. How is gene copy number determined?
- Q. Can PCR detect CNV?
- Q. What is transgene copy number?
- Q. How do you calculate copy number of plasmid?
- Q. What is copy number PCR?
- Q. How is virus copy number calculated?
- Q. How is quantitative PCR used to analyze gene expression?
- Q. What is the protocol for a qPCR thermocycle?
- Q. How does TaqMan work with real time PCR?
- Q. How to measure copy number of a gene?
Q. What is transgene copy number?
Transgene copy number is defined as the number of exogenous DNA insert(s) in the genome. For example, if the exogenous DNA fragment inserts only once at a single locus of the genome, it is a single copy transgenic event. The copy number is closely relevant to another concept, zygosity.
Q. How do you calculate copy number of plasmid?
Plasmid copy number was determined by comparing the quantification signal from the plasmid to those from the chromosome. Copy number was then calculated by using a known copy number plasmid as a standard.
Q. What is copy number PCR?
Precise genomic copy number variation analysis Copy number variation (CNV) is defined as a modification in the genome where the number of copies of a genomic DNA sequence differs from a reference or standard. CNVs are linked to susceptibility or resistance to disease, and thus are an important area for detailed study.
Q. How is virus copy number calculated?
The number of viral cDNA molecules in the standard sample was calculated using the formula: Y molecules μL-1=(X g μL-1 DNA/[base pairs of recombinant plasmid x 660]) x 6.022×1023 (QIAGEN, 2014.
Q. How is quantitative PCR used to analyze gene expression?
Quantitative PCR (qPCR) has been utilized for the analysis of gene expression (Heid et al., 1996; Higuchi et al., 1991) and quantification of copy number variation by real-time PCR. qPCR involves amplification of a test locus with unknown copy number and a reference locus with known copy number.
Q. What is the protocol for a qPCR thermocycle?
A typical qPCR experiment thermocycle protocol is as following, Cycle1 X 1 repeat Cycle2 X40 repeats Step1 Step 1 Step2 Temperature 95oC 95oC 60oC Dwell time 5min 0.5min 1min A typical qPCR experiment layout is as in the table below. There are 5 samples of serially diluted standard (S1, S2, S3, S4, and S5) for creating a standard curve, 2
Q. How does TaqMan work with real time PCR?
Applied Biosystems TaqMan Copy Number Assays combine TaqMan MGB probe chemistry with Applied Biosystems real-time PCR instruments to provide a method for obtaining specific, reproducible, and easy-to-interpret copy number results. This fast and simple method can be completed in hours rather than days.
Q. How to measure copy number of a gene?
This is a basic protocol for quantification of copy number from genomic DNA. To measure DNA copy number, the amplicon should be located either within an exon or intron with sequences unique to that gene. A control gene with two copies should also be included.